A World for Every Child
Capstone Overview
A World for Every Child aims to improve childhood cancer diagnosis in under-resourced areas using advanced machine learning. Focused on early and accurate detection, we developed two models to diagnose Acute Lymphoblastic Leukemia from blood smears. Despite the challenges of deploying such technology in developing nations, including resource limitations and the need for FDA clearance, our tool offers a quick, cost-effective preliminary diagnosis to aid doctors. This initiative aims to bridge the gap in global childhood cancer outcomes and introduces machine learning techniques to areas facing significant disparities in medical care.
Problem Motivation:
Global Inequalities in Childhood Cancer Outcomes
Each year, the World Health Organization reports that about 400,000 children globally are diagnosed with cancer. A child's chance of surviving cancer depends on their birthplace, with around 80% of children surviving in the Global North and less than 30% in the Global South. This disparity is due to several factors, such as access to medical care and accurate diagnosis. Particularly in developing regions, childhood cancers are often underdiagnosed.
We aim to enhance the early diagnosis rates of childhood cancer in underserved regions, with a specific emphasis on leukemia, a common form of cancer in children. Our efforts are focused on analyzing peripheral blood smears, a key diagnostic method for leukemia. Our target demographic is children in low-to-middle-income countries, as categorized by the World Bank.
Our problem is more than a classification problem. Deploying advanced image classification models for leukemia poses unique challenges. These models, which must accurately distinguish between different leukemia types, require advanced machine-learning techniques due to the complexity and ever-changing nature of the disease. Implementing these solutions in developing nations is difficult due to varied medical imaging technologies, limited resources like computing power and electricity, skilled labor shortages, and underdeveloped health infrastructure. Furthermore, obtaining mandatory FDA clearance, essential for ensuring safety and efficacy, adds significant time and cost to the process.
Solution: Boosting Diagnosis Rates Using Machine Learning
The core value proposition of our tool is to address and overcome several challenges commonly faced in developing regions. Traditional manual blood smear analysis, which can take hours or even days to process, is particularly challenging in resource-constrained areas. Additionally, financial constraints and scalability issues, coupled with the global disparities in medical care, further exacerbate the problem.
Using machine learning, we have developed two complementary models to diagnose childhood Acute Lymphoblastic Leukemia (ALL) from peripheral blood smears, enhancing early diagnosis and offering an alternative to conventional manual methods.
- Our tool aims to improve the manual process by providing an automated preliminary prediction in just a few seconds.
- A reduction in both time and cost for diagnoses, allows for an diagnosis of an estimated additional 100,000 children.
- Our tool is designed to augment the efforts of doctors in various regions, helping to bridge the diagnostic gaps and not replace their critical role in healthcare delivery.
Minimium Viable Product for Enhanced Diagnosis
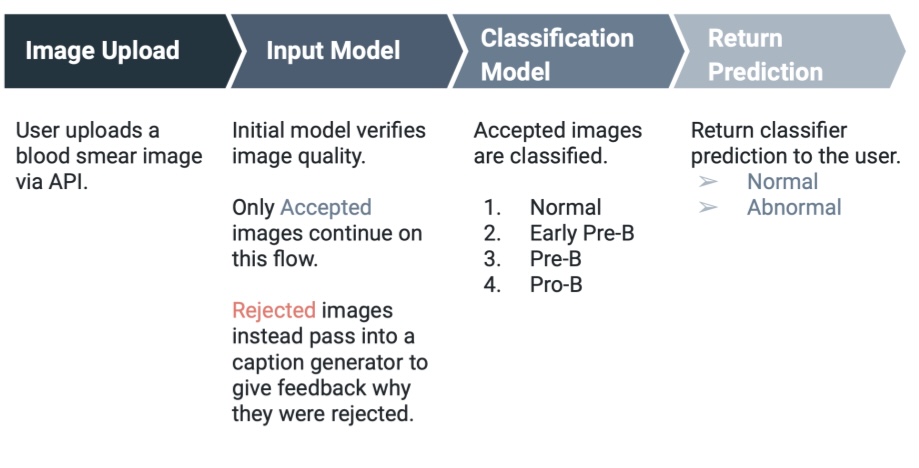
We have developed a three-part minimum viable product (MVP) for health professionals in under-resourced regions. This tool is designed to enhance diagnostic accuracy, alleviate diagnostic burden, and increase scalability, all at no cost to the user.
- Web application interface: where laboratory technicians can easily upload images and receive guidance on both uploading and interpreting the results from our image recognition system.
- Caption generation model: uses uploaded images to inform users about image quality, which is crucial for improving results, especially in areas with lower technological capabilities.
- Image recognition model: also uses the uploaded images. This model is crucial in increasing both the accuracy and speed of diagnosis, proving particularly beneficial in regions with a scarcity of skilled diagnostic personnel.
Data and Technical Details
Our project utilizes a dataset for Acute Lymphoblastic Leukemia image analysis, sourced from Taleqani Hospital in Tehran, Iran, and published on Kaggle. This dataset comprises 3,256 peripheral blood smear images from 89 patients, each labeled by a specialist, and is categorized into four distinct classes(normal, three malignant). Using this training data, we developed two complementary models to diagnose childhood ALL using PBS images.
1. Caption generation model approach
We chose the Generative Image-to-Text model due to its comprehensive, flexible, and scalable nature, which surpasses traditional image classifiers. This model allows for the easy integration of new alterations by simply defining new captions and pairing them with corresponding alteration functions.
For our main use case (good quality images), we achieved accurate captions for all 400 out of 400 high-quality images tested and the Word Score Error Rate (WER) rate was: 0.19 - 0.2 WER, classified as ‘Good to Excellent’.However, we acknowledge specific challenges and limitations as the model sometimes struggles with specific paired alterations, such as simultaneously decreasing brightness and contrast, often only effectively reducing brightness.
2. Image classification modeling approach
We opted for the EfficientNetB7 (ENB7) model developed by Google. This choice was driven by the model's significant accuracy and efficiency in handling various image classification tasks.It surpasses the accuracy and efficiency of other CNN models, making it highly effective for complex image classification tasks, such as leukemina classification. The model balances size, computational efficiency, and performance optimally. Its adaptability is particularly suited to leukemia's evolving nature, with algorithms capable of adjusting to the disease's changing characteristics. Additionally, its efficiency leads to faster training and inference times, which is advantageous in medical settings. The model's scalable architecture further ensures its adaptability to the varied nature of leukemia cases, making it an ideal choice for this challenging classification task.
The model excels at determining if a blood smear image is either normal or one of three types of malignant conditions, with accuracy between 97% and 99% in both validation and testing phases (within an acceptable range for medical applications).
However, the model faces challenges when dealing with images of low quality, often needing help to classify these accurately. Additionally, it's important to note that this model doesn't discriminate between relevant and irrelevant inputs, attempting to organize any image it receives, whether it's a blood smear or something completely unrelated. Lastly, the model is quite large, which might be a consideration for deployment, especially in environments with limited computing resources.
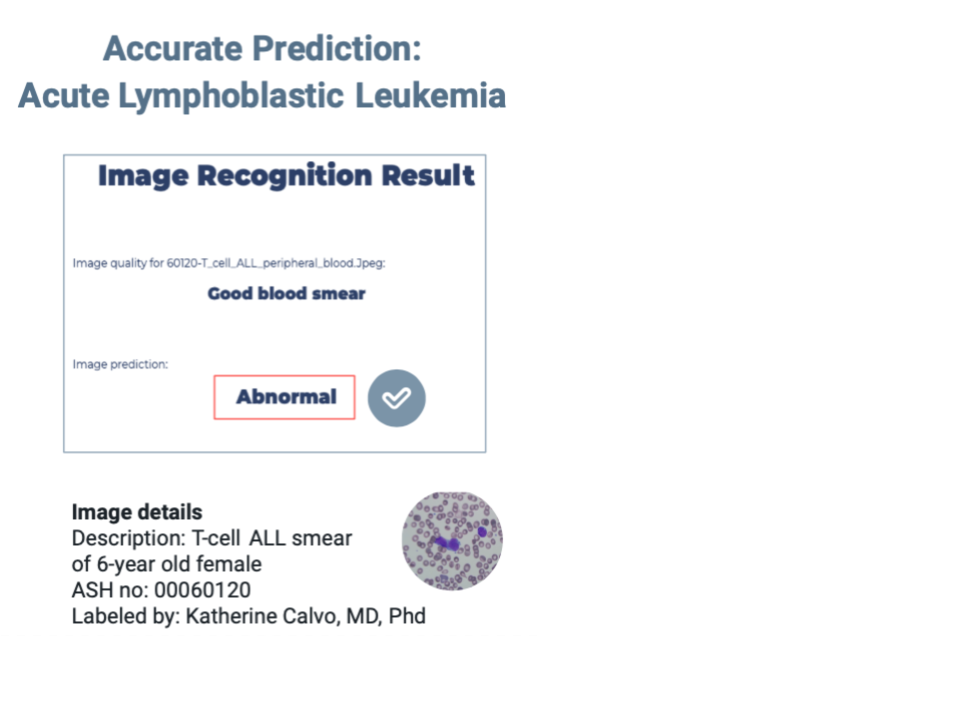
Promising Out-of-Sample Results
We tested our models using an open image dataset from the American Society of Hematology (ASH). This dataset is expertly annotated and aimed at helping new doctors prepare for their board exams. Although small (less than 25 images), our out-of-sample testing showed encouraging results. The model successfully identified a normal blood smear and an acute lymphoblastic leukemia blood smear from a 6-year-old patient. Interestingly, it correctly classified an acute myeloid leukemia image as abnormal (the training set was based on ALL images). However, it struggled with an ALL sample labelled with genetic irregularities. Many images did not meet our model's quality requirements, necessitating enhancements like magnification or contrast adjustments.
Ethical and Regulatory Considerations
Our project acknowledges the ethical responsibility of working with children, a vulnerable group, and we are committed to ensuring their data rights. A key element of our approach is informed consent, which involves explaining our tool to parents. Moreover, we are committed to rigorous data protection and security and emphasize the importance of data minimization, including anonymization.
FDA clearance is a requirement for deploying medical software, even if the product is intended for international use. This compliance is essential for ensuring the software's safety and efficacy but does increase the time, complexity and cost of the deployment process. Typically, models like these are classified as software as a Medical Device (SaMD), falling under Class II, denoting medium risk.
Key Learnings
We have much to learn from local medical providers to make our tool successful, including fully understanding the hurdles these regions face. Beyond providing a diagnostic tool, we hope that introducing advanced machine-learning techniques to these areas could lead to a virtuous cycle of technological advancement and better healthcare. To expand our project beyond this capstone, usability studies and partnerships with medical professionals will be critical, as it is obtaining FDA clearance, likely as Software as a Medical Device.
Acknowledgements
We thank our capstone supervisors, Korin Reid and Puya Vahabi, for their invaluable encouragement, insightful suggestions, and technical expertise throughout this project. We owe a debt of gratitude to Jared Maslin, for his extensive insights on privacy and regulatory considerations, including the FDA clearance process. Additionally, our appreciation goes to the external experts who deepened our understanding of deployment in developing economies with special thanks to Dr. Kelsey Brown, Ladi, Delano, and Rob Heath.